Introduction¶
Bibliometrics and semantic network¶
Bibliometrics contains a set of techniques that can help us manage and transform bibliographic information such as research papers to gain knowledge about a particular domain. It implies the use of statistical methods to assess what has been published about an specific topic, as well as the main authors, journals and countries of origin of the publications On the technical side it involves the manipulation of bibliographic reference files like .bib and graph extensions like .graphml or .gexf using software such as VosViewer, JabRef or Zotero.
The construction of semantic networks from a large corpus is another approach that can also provide insights in the form of trends and central terms in the area of study that is being targeted. These two approaches can throw light on emerging and novel fields of research that are useful to understand COVID-19. In our case, a combination of bibliometrics and network analysis is used to identify the relevant information that has been published related to vaccines.
A simple network mapping can reveal valuable insights into the development of SARS-CoV-2 vaccine even if you are not an specialist in medical research.
The filtering of information based on the insights provided by the networks led me to answer the following questions:
- Effectiveness of drugs being developed and tried to treat COVID-19 patients. 1.1. Clinical and bench trials to investigate less common viral inhibitors against COVID-19 such as naproxen, clarithromycin, and minocyclinethat that may exert effects on viral replication.
- Methods evaluating potential complication of Antibody-Dependent Enhancement (ADE) in vaccine recipients.
- Exploration of use of best animal models and their predictive value for a human vaccine.
- Capabilities to discover a therapeutic (not vaccine) for the disease, and clinical effectiveness studies to discover therapeutics, to include antiviral agents.
- Alternative models to aid decision makers in determining how to prioritize and distribute scarce, newly proven therapeutics as production ramps up. This could include identifying approaches for expanding production capacity to ensure equitable and timely distribution to populations in need.
- Efforts targeted at a universal coronavirus vaccine.
- Efforts to develop animal models and standardize challenge studies
- Efforts to develop prophylaxis clinical studies and prioritize in healthcare workers
- Approaches to evaluate risk for enhanced disease after vaccination
- Assays to evaluate vaccine immune response and process development for vaccines, alongside suitable animal models [in conjunction with therapeutics]
Data Exploration¶
The first stage is to view the .ris file that was built using the dataset provided in this challenge. The dataset is composed of around 32,000 research papers that relate to Covid-19. This information was used to create an undirected graph network with VosViewer, a science mapping software developed in Leiden University. This semantic network will include the most relevant terms based in binary counting, a method to count the presence of abscence of a term in the whole abstract or title being analyzed. When I create a map based on text data using our .ris file the following network is produced.

I will need to export this graph for further analysis in R. To do that, inside VosViewer I export the network as a graphML file. Fortunately VosViewer does the job of calculating each node weight, eigenvector centrality and modularity class, measures that will be useful to identify the most relevant terms in the dataset as well as the cluster they belong to.
#install.packages("igraph")
library(igraph)
g1 <- read.graph("https://raw.githubusercontent.com/mdurazob/covidnotebook/master/AbstractsCovid19.graphml", format="graphml")
library(igraph)
V(g1)
We need to extract the nodes that are related to "vaccine strain" and "vaccine candidate". (We can see all the n-grams that contain the word vaccine in the filter section of VosViewer or by using the igraph package)
library(igraph)
nodesofinterest <- c("therapeutic vaccine", "vaccine candidate")
# select the nodes having these names
nodesselection <- V(g1)[label %in% nodesofinterest]
# get their neighborhood
egoneighborhood <- ego(g1, order=1, nodes = nodesselection, mode = "all", mindist = 0)
# turn the returned list into a graph
egoselection <- induced_subgraph(g1,unlist(egoneighborhood))
V(egoselection)
The resulting graph contains 739 nodes or vertices. We will refine this result further by plotting only those nodes whose links weight is above 1000, and use the fruchtermain-reingold force directed algorithm to produce a layout to host the nodes. The force between nodes defines their place in the network.
This algorithm guarantees that topologically near nodes are placed in the same vicinity, and far nodes are placed far from each other. This way, nodes in a vicinity have stronger relations between them than what they have with nodes in another vicinity.
#install.packages("GGally")
library(GGally)
#install.packages("sna")
library(sna)
library(network)
install.packages("intergraph")
install.packages("ggnet2")
library(ggplot2)
ggnet2(egoselection, mode = 'fruchtermanreingold', node.size="weightlinks", node.color = "cluster", label = "label", label.size=2.5, palette="Dark2", alpha = 0.75, edge.alpha = 0.2, size.legend = NULL, size.min = 1000) + theme(legend.position = "none")
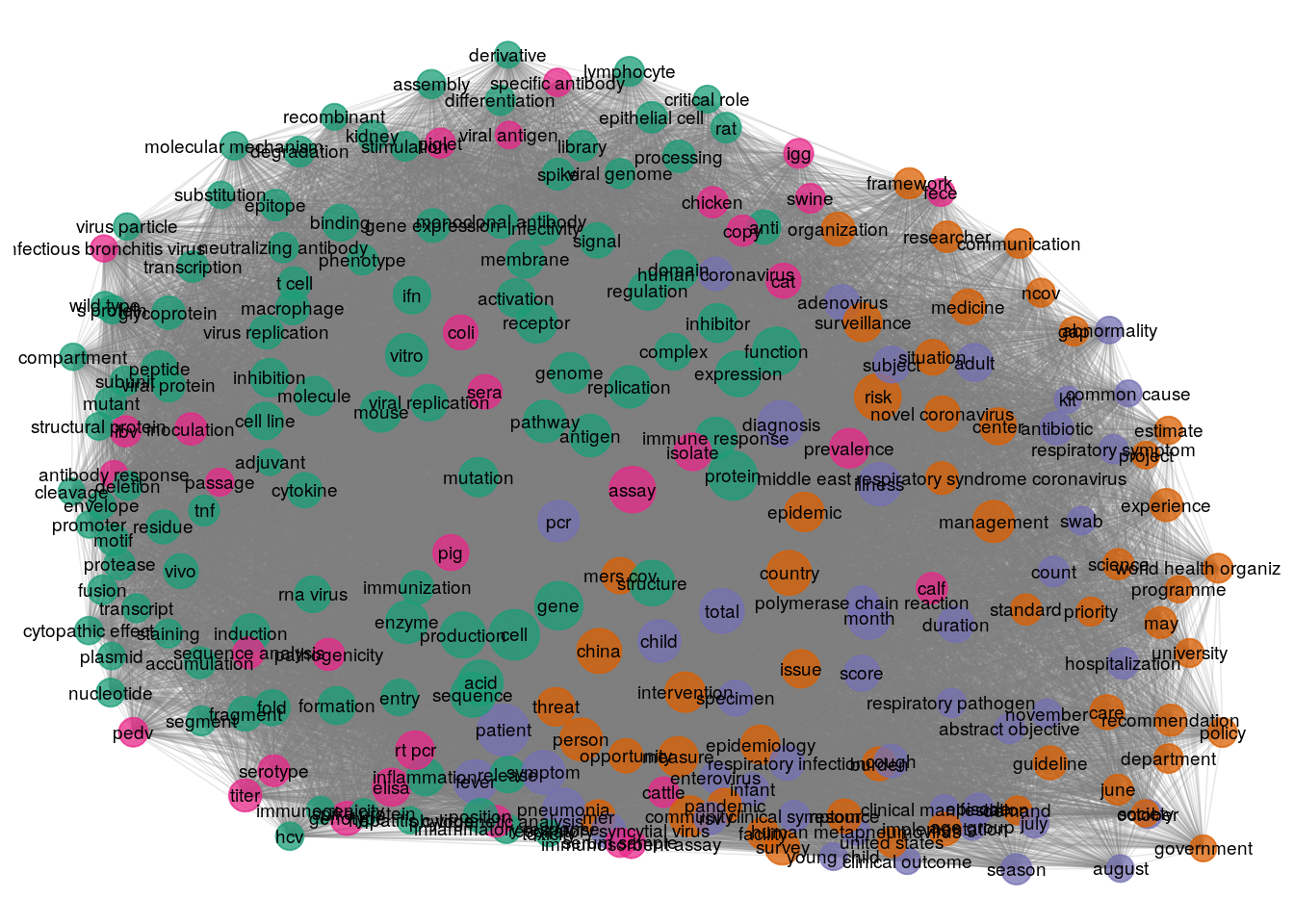
Each color in this graph represents a cluster. Nodes colored the same way have stronger connections between them than what they have with other nodes of another color. In this case, the nodes in purple color carry a meaning that is related to methodological and scientific terminology, these nodes are the ones that can provide insights and valuable knowledge to answer the questions provided in this challenge.
Side note: We can also add interactivity to our graph by saving our filtered graph in a new graphML file and exporting it to a gexf file using Gephi. This gexf can then be plotted in R using simajs package. If we follow this steps, we can add the ability to zoom in and out by using our gexf file.
#install.packages("sigmajs")
library(sigmajs)
reducedgraph <- "https://raw.githubusercontent.com/mdurazob/covidnotebook/master/VaccineGraphReduced.gexf"
vaccineInteractive <- sigmajs() %>%
sg_from_gexf(reducedgraph) %>%
sg_settings(drawLabels = TRUE, drawEdges = FALSE, labelThreshold = 4, labelSize = 10)
vaccineInteractive
library(sigmajs)
cluster1graph <- "https://raw.githubusercontent.com/mdurazob/covidnotebook/master/Cluster1.gexf"
cluster1interactive <- sigmajs() %>%
sg_from_gexf(cluster1graph) %>%
sg_settings(drawLabels = TRUE, drawEdges = FALSE, labelThreshold = 4, labelSize = 10)
cluster1interactive
To further explore the terms in this cluster, we can visualize the edges coming to and from them using a Sankey Diagram.
The sankey diagram can summarize the data inside the network. However it can leave out meaningful terms that do not have enough links to be relevant. Some of the terms highlighted in this chart are: spike protein, t cell, s protein, t lymphocite, among others.
#install.packages("networkD3")
## Installing package into '/usr/local/lib/R/site-library'
## (as 'lib' is unspecified)
library(networkD3)
edges <- read.csv(file ="https://raw.githubusercontent.com/mdurazob/covidnotebook/master/edgescluster1.csv")
nodes <- read.csv(file = "https://raw.githubusercontent.com/mdurazob/covidnotebook/master/nodescluster1.csv")
d <- sankeyNetwork(Links = edges, Nodes = nodes, Source = "Source", Target = "Target",
NodeID = "name", Value = "Weight", # LinkGroup = "colour", (use if you want to colour edges)
fontSize = 14, fontFamily = "sans-serif", width = "100%")
d
The following are some definitions that can help us to better understand the field that we are examining.
The spike protein, a main surface antigen of SARS-CoV, is one of the most important antigen candidates for vaccine design. The spike protein (S) of the virus enables binding to the human receptor ACE2, the S protein represents a prime target for vaccines preventing viral entry into human host cells.
The S proteins from SARS-CoV-1 and SARS-CoV-2 are similar, but structural differences in the receptor binding domain (RBD) preclude the use of SARS-CoV-1–specific neutralizing antibodies to inhibit SARS-CoV-2.
Recent research by Jason S. McLellan of UTAustin reveals the 3D structure of this spike protein by using cryoSPARC, a novel biotech software to model biological structures. The precision and resolution can pave the way for scientists to identify the differences between SARS-CoV1 and SARS-CoV2 with the aim of developing a vaccine of the latter.

Other terms highlighted are T cell and T lymphocite. A quick search revealed that currently some vaccine developments focus on T cell therapy:
Now, AlloVir and Baylor are applying the same approach to the SARS-CoV-2 virus sweeping across the world. AlloVir is aiming to create an off-the-shelf therapy that is capable of targeting SARS-CoV-2 and potentially similar viruses such as SARS-CoV, MERS-CoV and endemic coronaviruses.
The plan is to position the SARS-CoV-2-specific T cells for use as a monotherapy and incorporate the coronaviruses into ALVR106, a preclinical asset aimed at community-acquired respiratory viruses. In doing so, AlloVir thinks it can improve outcomes in immunocompromised patients who are exposed to SARS-CoV-2 and other viruses with the potential to overwhelm their weakened immune systems.
As we can see there is some relation shared by the highlighted terms and the state of the art on the development of the vaccine. The current efforts to develop the vaccine are directly linked to the terms highlighted in the previous analysis.
We will use these terms to look for research papers that mention them in their title or abstract along a keyword selected from each of the questions of this challenge.
Answers to task questions¶
Effectiveness of drugs being developed and tried to treat COVID-19 patients.¶
Clinical and bench trials to investigate less common viral inhibitors against COVID-19 such as naproxen, clarithromycin, and minocyclinethat that may exert effects on viral replication.¶
Recent research has unveiled that α-Ketoamide inhibitors can target the viral main protease Mpro, also called 3CLpro. Other research suggest ACE-2 based peptides, remdesivir and a novel vinylsulfone can be succesful protease inhibitors. The following chart illustrates the most relevant research papers including potential inhibitors, size indicates the number of google scholar citations.
Umifenovir is a promising repurposed antiviral agent with a unique mechanism of action targeting the S protein/ACE2 interaction and inhibiting membrane fusion of the viral envelope.
Other immunomodulatory agents as baricitinib, imatinib, dasatinib, and cyclosporine demonstrate in vitro activity purported to inhibit SARS-CoV-2.
Nitazoxanide also has demonstrated in vitro antiviral activity against MERS and SARS-CoV-2
Favipiravir was compared with umifenovir (also known as Arbidol) for the treatment of SARS-CoV-2. In terms of statistical relevance, the study highlights this results:
Differences in clinical recovery at day 7 were observed in patients with moderate infections (71.4% favipiravir and 55.9% Arbidol, P = .019).
#install.packages("highcharter")
library(highcharter)
inhibitionpapers <- read.csv("https://raw.githubusercontent.com/mdurazob/covidnotebook/master/InhibitionPapers.csv")
inhibitionchart <- inhibitionpapers %>%
hchart('bubble', hcaes(x = 'Name', y = 'Year', size = 'Citations', color='Year')) %>%
hc_tooltip(pointFormat = "<b>Title: {point.Title}</b><br> <b>Keyword: {point.Name}</b><br> Fragment: {point.Abstract}")
inhibitionchart
| Year | Name | Title | Abstract | Citations |
|---|---|---|---|---|
| 2020 | Remdesivir | Learning from the Past: Possible Urgent Prevention and Treatment Options for Severe Acute Respiratory Infections Caused by 2019-nCoV | Herein, we suggest 4 potential drug candidates (an ACE2-based peptide, remdesivir, 3CLpro-1 and a novel vinylsulfone protease inhibitor) that can be used to treat patients suffering with the 2019-nCoV. | 30 |
| 2020 | Favipiravir/Remdesivir | Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. | In this study, we evaluated the antiviral efficiency of five FAD-approved drugs including ribavirin, penciclovir, nitazoxanide, nafamostat, chloroquine and two well-known broad-spectrum antiviral drugs remdesivir (GS-5734) and favipiravir (T-705) against a clinical isolate of 2019-nCoV in vitro. | 384 |
| 2018 | TMPRSS11A | TMPRSS11A activates the influenza A virus hemagglutinin and the MERS coronavirus spike protein and is insensitive against blockade by HAI-1 | Our results suggest that TMPRSS11A could promote FLUAV spread in target cells and that HA-activating TTSPs exhibit differential sensitivity to blockade by cellular serine protease inhibitors. | 3 |
| 2018 | IFITM2 and IFITM3 | Identification of Residues Controlling Restriction versus Enhancing Activities of IFITM Proteins on Entry of Human Coronaviruses | IFITM3 reduced its activity to inhibit the entry of HCoV-NL63 and -229E but enhanced the entry of SARS-CoV and MERS-CoV. Second, replacement of IFITM3 tyrosine 99 with either alanine or aspartic acid reduced its activity to inhibit the entry of HCoV-NL63 and SARS-CoV but promoted the entry of MERS-CoV | 15 |
| 2018 | 229E-HR2P | Peptide-Based Membrane Fusion Inhibitors Targeting HCoV-229E Spike Protein HR1 and HR2 Domains | We found that 229E-HR1P and 229E-HR2P could interact to form a stable six-helix bundle and inhibit HCoV-229E spike protein-mediated cell-cell fusion with IC50 of 5.7 and 0.3 µM, respectively. 229E-HR2P effectively inhibited pseudotyped and live HCoV-229E infection with IC50 of 0.5 and 1.7 µM, respectively. | 16 |
| 2017 | Umifenovir (Arbidol) | Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. | This unique binding mode compared with the small-molecule inhibitors of other class I fusion proteins enhances our understanding of how small molecules can function as fusion inhibitors and guides the development of broad-spectrum therapeutics against influenza virus. | 0 |
| 2017 | 5F9 mAb | A novel neutralizing monoclonal antibody targeting the N-terminal domain of the MERS-CoV spike protein. | In addition to mAbs with neutralizing epitopes located on the receptor-binding domain, one mAb, 5F9, which binds to the N-terminal domain (NTD) of the MERS-CoV S1 subunit, showed efficient neutralizing activity against the wild-type MERS-CoV strain EMC/2012, with a half maximal inhibitory concentration of 0.2??g/mL | 21 |
| 2016 | Nitazoxanide | Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus | Nitazoxanide is a broad-spectrum antiviral agent undergoing clinical development for treatment of influenza and other viral respiratory infections. Nitazoxanide exhibits in vitro activity against Middle East respiratory syndrome coronavirus (MERS-CoV) and other coronaviruses, inhibiting expression of the viral N protein. Nitazoxanide also suppresses production of pro-inflammatory cytokines in peripheral blood mononuclear cells and suppresses interleukin 6 production in mice | 26 |
| 2016 | S2 Subunit | Inhibition of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) infectivity by peptides analogous to the viral spike protein. | The Wimley and White interfacial hydrophobicity scale was used to identify regions within the CoV S2 subunit that may preferentially associate with lipid membranes with the premise that peptides analogous to these regions may function as inhibitors of viral infectivity. | 57 |
| 2015 | Compound 5 | Three new coumarins from Saposhnikovia divaricata and their porcine epidemic diarrhea virus (PEDV) inhibitory activity | Among all isolates, compound 5 revealed strongest inhibitory effect on PEDV replication. Quantitative real-time PCR data showed inhibitory effect of 5 on genes responsible for synthesis of PEDV vital structural proteins (GP6 nucleocapsid, GP2 spike, and GP5 membrane) in a dose-dependent manner. Also, compound 5 demonstrated the inhibitory effect on PEDV GP6 nucleocapsid and GP2 spike protein synthesis as analyzed by western blotting. | 14 |
| 2014 | S1 Subunit | Middle East respiratory syndrome coronavirus (MERS-CoV) entry inhibitors targeting spike protein. | Here we discuss the most recent studies on the structure of the MERS-CoV spike protein and its role in virus binding and entry, and the development of MERS-CoV entry/fusion inhibitors targeting the S1 subunit, particularly the receptor-binding domain (RBD), and the S2 subunit, especially the HR1 region, of the MERS-CoV spike protein. | 45 |
| 2014 | pAPN | A phage-displayed peptide recognizing porcine aminopeptidase N is a potent small molecule inhibitor of PEDV entry | Three phage-displayed peptides designated H, S and F that recognize porcine aminopeptidase N (pAPN), the cellular receptor of porcine transmissible gastroenteritis virus (TGEV) were able to inhibit cell infection by TGEV | 14 |
| 2013 | NH4Cl and bafilomyci | Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. | Surprisingly, a polyclonal antibody to the S protein of MHV, a group a murine betacoronavirus, cross-reacted in immunoblots with the S2 domain of group c MERS-CoV spike protein. MERS pseudovirions released from 293T cells contained only uncleaved S, and pseudovirus entry was blocked by lysosomotropic reagents NH4Cl and bafilomycin and inhibitors of cathepsin L. | 110 |
| 2012 | Tyr-Lys-Tyr-Arg-Tyr-Leu Hexapeptide | A hexapeptide of the receptor-binding domain of SARS corona virus spike protein blocks viral entry into host cells via the human receptor ACE2. | In vitro infection of Vero E6 cells by SARS coronavirus (SARS-CoV) is blocked by hexapeptide Tyr-Lys-Tyr-Arg-Tyr-Leu. | 17 |
| 2011 | Cyclosporin A | Cyclosporin A inhibits the replication of diverse coronaviruses. | Upon high-multiplicity infection, CsA treatment rendered SARS-CoV RNA and protein synthesis almost undetectable, suggesting an early block in replication. | 60 |
| 2011 | LF | Inhibition of SARS Pseudovirus Cell Entry by Lactoferrin Binding to Heparan Sulfate Proteoglycans | We further investigated the role of LF in the entry of SARS pseudovirus into HEK293E/ACE2-Myc cells. Our results reveal that LF inhibits SARS pseudovirus infection in a dose-dependent manner | 25 |
| 2010 | TACE | TACE antagonists blocking ACE2 shedding caused by the spike protein of SARS-CoV are candidate antiviral compounds. | Because outbreaks of severe acute respiratory syndrome coronavirus (SARS-CoV) might reemerge, identifying antiviral compounds is of key importance. Previously, we showed that the cellular factor TNF-alpha converting enzyme (TACE), activated by the spike protein of SARS-CoV (SARS-S protein), was positively involved in viral entry, implying that TACE is a possible target for developing antiviral compounds. | 24 |
| 2009 | PBMC | SARS coronavirus spike protein-induced innate immune response occurs via activation of the NF-?B pathway in human monocyte macrophages in vitro | A purified recombinant spike (S) protein was studied for its effect on stimulating human peripheral blood monocyte macrophages (PBMC). We examined inflammatory gene mRNA abundances found in S protein-treated PBMC using gene arrays. | 33 |
| 2009 | Enediynyl peptide | A novel enediynyl peptide inhibitor of furin that blocks processing of proPDGF-A, B and proVEGF-C | Circular dichroism study showed that this inhibitor displayed a predominantly beta-turn structure while western blots confirmed its ability to protect furin protein from self degradation. These findings imply its potential as a therapeutic agent for intervention of cancer and other furin-associated diseases. | 23 |
| 2007 | Ben-HCl | Cleavage of spike protein of SARS coronavirus by protease factor Xa is associated with viral infectivity | However, other functions of SARS-CoV S protein such as proteolytic cleavage and its implications to viral infection are incompletely understood. In this study, we demonstrated that the infection of SARS-CoV and a pseudovirus bearing the S protein of SARS-CoV was inhibited by a protease inhibitor Ben-HCl. | 49 |
| 2006 | SP4, SP8 and SP10 | Design and biological activities of novel inhibitory peptides for SARS-CoV spike protein and angiotensin-converting enzyme 2 interaction. | Therefore, we assayed the inhibitory effects of small peptides derived from S protein on the binding of S protein to ACE2 and on the S-protein-pseudotyped retrovirus infectivity. SP-4 (residues 192-203), SP-8 (residues 483-494), and SP-10 (residues 668-679) significantly blocked the interaction between S protein and ACE2 by biotinylated enzyme-linked immunosorbent assay, with IC(50) values of 4.30 +/- 2.18, 6.99 +/- 0.71, and 1.88 +/- 0.52 nmol, respectively. | 27 |
| 2005 | HR121 | Design of recombinant protein-based SARS-CoV entry inhibitors targeting the heptad-repeat regions of the spike protein S2 domain. | However, synthesis of these peptides is at high cost. In this study, we designed two recombinant proteins, one containing two HR1 and one HR2 peptides (denoted HR121), and the other consisting of two HR2 and one HR1 peptides (designated HR212). These two proteins could be easily purified with the low cost of production, exhibiting high stability and potent inhibitory activity on entry of the HIV/SARS pseudoviruses with IC(50) values of 4.13 and 0.95muM, respectively. | 30 |
Methods evaluating potential complication of Antibody-Dependent Enhancement (ADE) in vaccine recipients.¶
With the emerging development of monoclonal antibodies as therapeutics, the potential implications of antibody-dependent enhancement of disease need to be addressed.
Some investigations suggest that generating monoclonal antibodies against SARS-CoV spike proteins may trigger ADE effects.
One solution suggested in a 2016 study asserts that peptide-based vaccines against SARS-CoV could be engineered to avoid ADE via elimination of the S597-603 epitope.
A more recent paper published in March 2020 shows small bit of cross-neutral activity between original SARS-CoV and SARS-Cov2 by analyzing plasma samples. The study finds that while cross-reactivity in antibody binding to the spike protein is common, cross-neutralization of the live viruses is rare, indicating the presence of non-neutralizing antibody response to conserved epitopes in the spike.
Other particular study assesed a recombinant native full length S-protein trimer (triSpike) to elicit an immune response while analyzing the capacity of anti-S antibodies to mediate ADE of virus entry.
Another study found that antibody-mediated infection was dependent on signaling-competent members of the human FcγRII family, which were shown to confer susceptibility to otherwise naïve ST486 cells, as binding of immune complexes to cell surface FcγRII was necessary but not sufficient to trigger antibody-dependent enhancement (ADE) of infection.
In a similar fashion this article confirms that antibody mediated infection is indeed dependent on FcYRII but does not use the endosomal/lysosomal pathway utilized by angiotensin I converting enzyme 2 (ACE2), the accepted receptor for SARS-CoV.This suggests that ADE of SARS-CoV utilizes a novel cell entry mechanism into immune cells.
Taking a different approach this article suggests that low levels of neutralizing antibodies in SARS-CoV Z-1 vaccine immunized Chinese rhesus macaques, hematological assay results were in the normal range, and no elevation of biochemical indices was observed after immunization, the scientists claim that this model could be used for testing ADE of the vaccine candidate.
library(highcharter)
#install.packages("treemap")
library(treemap)
#install.packages("viridis")
library(viridis)
#install.packages("RColorBrewer")
library(RColorBrewer)
adepapers <- read.csv("https://raw.githubusercontent.com/mdurazob/covidnotebook/master/ADEPapers.csv")
hchart(adepapers, "treemap", hcaes(x = Name, value = Citations, color = Year))%>%
hc_tooltip(pointFormat = "<b>Title: {point.Title}</b><br> <b>Keyword: {point.Name}</b><br> Fragment: {point.Abstract}") %>%
hc_colorAxis(minColor = brewer.pal(9, "Greens")[4],
maxColor = brewer.pal(9, "Blues")[9])
| Year | Name | Author | Title | Abstract | Citations |
|---|---|---|---|---|---|
| 2020 | cross-reactivity | Huibin Lv | Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections | Our results show that while cross-reactivity in antibody binding to the spike protein is common, cross-neutralization of the live viruses is rare, indicating the presence of non-neutralizing antibody response to conserved epitopes in the spike. | 0 |
| 2018 | Low level SARS-CoV Z-1 | F Luo | Evaluation of Antibody-Dependent Enhancement of SARS-CoV Infection in Rhesus Macaques Immunized with an Inactivated SARS-CoV Vaccine | The results suggested that the inactivated vaccine was safe and the immunized rhesus macaques could be used as the model for testing ADE of the candidate vaccine. | 2 |
| 2016 | Tamiflu & M2 | McKimm Breschkin | Meeting report: 4th ISIRV antiviral group conference: Novel antiviral therapies for influenza and other respiratory viruses | With the emerging development of monoclonal antibodies as therapeutics, the potential implications of antibody-dependent enhancement of disease were also addressed. Topics covered all aspects from structural and molecular biology to preclinical and clinical studies. | 34 |
| 2016 | S597-603 | Wang Q | Immunodominant SARS Coronavirus Epitopes in Humans Elicited both Enhancing and Neutralizing Effects on Infection in Non-human Primates | Thus, peptide-based vaccines against SARS-CoV could be engineered to avoid ADE via elimination of the S597-603 epitope. We provide herein an alternative strategy to prepare a safe and effective vaccine for ADE of viral infection by identifying and eliminating epitope sequence-dependent enhancement of viral infection. | 10 |
| 2014 | Monoclonal antibodies | SF Wang | Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins | Results from infectivity assays indicate that SARS-CoV ADE is primarily mediated by diluted antibodies against envelope spike proteins rather than nucleocapsid proteins. We also generated monoclonal antibodies against SARS-CoV spike proteins and observed that most of them promoted SARS-CoV infection. Combined, our results suggest that antibodies against SARS-CoV spike proteins may trigger ADE effects. | 37 |
| 2014 | FcyRII | Yip MS | Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus | These results demonstrate that human macrophages can be infected by SARS-CoV as a result of IgG-mediated ADE and indicate that this infection route requires signaling pathways activated downstream of binding to Fc?RII receptors. | 23 |
| 2012 | IFITM Protein | YK Chan | IFITM Proteins Restrict Antibody-Dependent Enhancement of Dengue Virus Infection | We observed that IFITM proteins restricted ADE-mediated and direct infection with comparable efficiencies in a myelogenous leukemia cell line. Our data suggest that IFITM proteins can contribute to control of secondary DENV infections. | 42 |
| 2011 | Raji B cell | MS Yip | Investigation of Antibody-Dependent Enhancement (ADE) of SARS coronavirus infection and its role in pathogenesis of SARS | The occurrence of antibody-mediated infection of human Raji B cells was further investigated by using live SARS-CoV. Similarly to results obtained with the SARS-CoVpp, only anti-SARS-CoV Spike serum, but not mock immune-serum, induced a massive increase of SARS-CoV viral genes (ORF1b and Nucleocapsid) and viral proteins (Membrane and Nucleocapsid) in Raji B cells. | 1 |
| 2011 | FcyRII | M Jaume | Anti-Severe Acute Respiratory Syndrome Coronavirus Spike Antibodies Trigger Infection of Human Immune Cells via a pH- and Cysteine Protease-Independent Fc?R Pathway | Antibody-mediated infection was dependent on Fc? receptor II but did not use the endosomal/lysosomal pathway utilized by angiotensin I converting enzyme 2 (ACE2), the accepted receptor for SARS-CoV. This suggests that ADE of SARS-CoV utilizes a novel cell entry mechanism into immune cells. | 36 |
| 2007 | TNF-alpha | T Takano | TNF-alpha, produced by feline infectious peritonitis virus (FIPV)-infected macrophages, upregulates expression of type II FIPV receptor feline aminopeptidase N in feline macrophages | The pathogenicity of feline infectious peritonitis virus (FIPV) is known to depend on macrophage tropism, and this macrophage infection is enhanced by mediation via anti-S antibody (antibody-dependent enhancement, ADE). In this study, we found that TNF-alpha production was increased with viral replication in macrophages inoculated with a mixture of FIPV and anti-S antibody, and demonstrated that this culture supernatant had feline PBMC apoptosis-inducing activity | 52 |
| 2007 | triSpike S Protein trimer | YW Kam | Antibodies against trimeric S glycoprotein protect hamsters against SARS-CoV challenge despite their capacity to mediate Fc?RII-dependent entry into B cells in vitro | We investigated whether a recombinant native full-length S-protein trimer (triSpike) of severe acute respiratory syndrome coronavirus (SARS-CoV) was able to elicit a neutralizing and protective immune response in animals and analyzed the capacity of anti-S antibodies to mediate antibody-dependent enhancement (ADE) of virus entry in vitro and enhancement of replication in vivo. | 58 |
| 1992 | Monoclonal antibodies | Corapi VW | Monoclonal antibody analysis of neutralization and antibody-dependent enhancement of feline infectious peritonitis virus. | In most instances, neutralizing MAbs were able to induce ADE, demonstrating a direct relationship between neutralization and enhancement. The difference in immunoglobulin subclass between neutralizing MAbs that induced ADE and those that did not induce ADE suggests that there may be a restriction in the immunoglobulin subclasses capable of mediating ADE. | 114 |
| 1991 | M Protein | H Vennema | Primary structure of the membrane and nucleocapsid protein genes of feline infectious peritonitis virus and immunogenicity of recombinant vaccinia viruses in kittens | Feline infectious peritonitis virus (FIPV) causes a mostly fatal, immunologically mediated disease in cats. Previously, we demonstrated that immunization with a recombinant vaccinia virus expressing the FIPV spike protein (S) induced early death after challenge with FIPV (Vennema et al., 1990, J. Virol. 64, 1407-1409). In this paper we describe similar immunizations with the FIPV membrane (M) and nucleocapsid (N) proteins. The genes encoding these proteins were cloned and sequenced. Comparison of the amino acid sequences with the corresponding sequences of porcine transmissible gastroenteritis virus revealed 84.7 and 77% identity for M and N, respectively. Vaccinia virus recombinants expressing the cloned genes induced antibodies in immunized kittens. Immunization with neither recombinant induced early death after challenge with FIPV, strongly suggesting that antibody-dependent enhancement is mediated by antibodies against S only. Immunization with the N protein recombinant had no apparent effect on the outcome of challenge. However, three of eight kittens immunized with the M protein recombinant survived the challenge, as compared to one of eight kittens of the control group. | 83 |
Exploration of use of best animal models and their predictive value for a human vaccine.¶
A summary of a 2005 technical meeting in London summarizes the animal models and antibody assays for evaluating candidate SARS vaccines, the study highlights that:
In selecting animal models for vaccine evaluation, it is important to remember the principle underlying the so called “animal rule”, where data from more than one animal species is often required: each animal species should contribute something different to our understanding of disease and protection.
Upon this guideline the recommended animal models include: Inbred mouse strains (BALB/c, C57BL/6, 129SvEv, STAT1−/−) whom support SARS-CoV replication and can develop pneumonitis (129S). Golden Syrian hamsters are also an excellent animal model because they demonstrate high levels of SARS-CoV replication and develop pneumonitis. Hamsters are suitable for vaccine efficacy, immunoprophylaxis and treatment studies.
Among non-human primates (NHP) are the common marmoset and African green monkeys (AGMs, Chlorocebus aethiops sabeus).
The SARSCoV strains tested in NHP models are HKU-39849 (cynos), PUMC (rhesus) and Urbani (marmosets and AGMs). These were inoculated by the respiratory route (IN, IT) at a dose of 106pfu or more/NHP.
Another article analyzes the advantages and disadvantages of commonly used animals used in highly pathogenic disease models (members of the Filoviridae, Arenaviridae, Bunyaviridae and Paramyoviridae).
This chart summarizes the animal models present in research articles that realte to SARS-CoV and SARS-CoV2
library(highcharter)
animalmodelspapers <- read.csv("https://raw.githubusercontent.com/mdurazob/covidnotebook/master/AnimalModelsPapers.csv")
animalmodelschart <- animalmodelspapers %>%
hchart('bubble', hcaes(x = 'Name', y = 'Year', size = 'Citations', color='Year')) %>%
hc_tooltip(pointFormat = "<b>Title: {point.Title}</b><br> <b>Keyword: {point.Name}</b><br> Fragment: {point.Abstract}")
animalmodelschart
| Year | Name | Author | Title | Abstract | Citations |
|---|---|---|---|---|---|
| 1990 | Mice | Daniel C | Protection from lethal coronavirus infection by affinity-purified spike glycoprotein of murine hepatitis virus, strain A59 | Murine hepatitis viruses provide excellent animal models for the study of virus-induced diseases of the central nervous system and gastrointestinal tract. Several studies have indirectly provided evidence that the spike glycoprotein (S) of these coronaviruses bears determinants for pathogenesis and the induction of protective immunity. | 44 |
| 2005 | Murinae | Zhi Y | Identification of murine CD8 T cell epitopes in codon-optimized SARS-associated coronavirus spike protein | The causative agent of severe acute respiratory syndrome (SARS) has been identified as a new type of coronavirus, SARS-associated coronavirus (SARS-CoV). CD8 T cells play an important role in controlling diseases caused by other coronaviruses and in mediating vaccine-induced protective immunity in corresponding animal models. | 43 |
| 2005 | Immunological | Roberts A | Animal models and antibody assays for evaluating candidate SARS vaccines: Summary of a technical meeting 25–26 August 2005, London, UK | Administrations "animal rule" would apply to licensure of a SARS vaccine, it is important to develop standardized animal models and immunological assays in preparation for this eventuality. | 15 |
| 2007 | Mice | Du L | Receptor-binding domain of SARS-CoV spike protein induces long-term protective immunity in an animal model | In this study, BALB/c mice were vaccinated intramuscularly (i.m.) with 10microg of RBD-Fc (RBD fused with human IgG1 Fc) and boosted twice at 3-week intervals and one more time at 12th month. Humoral immune responses of vaccinated mice were investigated for up to 12 months at a 1-month interval and the neutralizing titers of produced antibodies were reported at months 0, 3, 6 and 12 post-vaccination. | 62 |
| 2007 | F344 Rats | J Virol | Participation of both Host and Virus Factors in Induction of Severe Acute Respiratory Syndrome (SARS) in F344 Rats Infected with SARS Coronavirus | By contrast, adult rats infected with the passaged virus showed respiratory symptoms and severe pathological lesions in the lung. Levels of inflammatory cytokines in sera and lung tissues were significantly higher in adult F344 rats than in young rats. During in vivo passage of SARS-CoV, a single amino acid substitution was introduced within the binding domain of the viral spike protein recognizing angiotensin-converting enzyme 2 (ACE2), which is known as a SARS-CoV receptor. T | 33 |
| 2007 | Mice | Barry Rockx | Synthetic Reconstruction of Zoonotic and Early Human Severe Acute Respiratory Syndrome Coronavirus Isolates That Produce Fatal Disease in Aged Mice | Using synthetic biology and reverse genetics, we describe the construction of a panel of isogenic SARS coronavirus (SARS-CoV) strains bearing variant spike glycoproteins that are representative of zoonotic strains found in palm civets and raccoon dogs, as well as isolates spanning the early, middle, and late phases of the SARS-CoV epidemic | 57 |
| 2013 | Multiple | D Safronetz | Animal models for highly pathogenic emerging viruses | For these reasons it is imperative that animal models of disease recapitulate the human condition as closely as possible in order to provide the best predictive data with respect to the potential efficacy in humans. In this article we review the current status of disease models for highly pathogenic and emerging viral pathogens. | 23 |
| 2014 | Mice | Agnihothram S | A Mouse Model for Betacoronavirus Subgroup 2c Using a Bat Coronavirus Strain HKU5 Variant | We use this platform to (i) identify a broad-spectrum antiviral that can potentially inhibit viruses closely related to MERS-CoV, (ii) demonstrate the absence of increased eosinophilic immune pathology for MERS-CoV N protein-based vaccines, and (iii) mouse adapt this virus to identify viral genetic determinants of cross-species transmission and virulence. This study holds significance as a strategy to control newly emerging viruses. | 31 |
| 2015 | Ferret | DY Oh | A Novel Video Tracking Method to Evaluate the Effect of Influenza Infection and Antiviral Treatment on Ferret Activity | Ferrets are the preferred animal model to assess influenza virus infection, virulence and transmission as they display similar clinical symptoms and pathogenesis to those of humans. | 18 |
| 2015 | Cross-Mouse | LE Gralinski | Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross | Using genetically diverse animals from incipient lines of the Collaborative Cross mouse panel, we demonstrate a greatly expanded range of phenotypes relative to classical mouse models of SARS-CoV infection including lung pathology, weight loss and viral titer. | 69 |
| 2017 | Ferret | Layton DS | Development of an anti-ferret CD4 monoclonal antibody for the characterisation of ferret T lymphocytes | The ferret is an established animal model for a number of human respiratory viral infections, such as influenza virus and more recently, Ebola virus. However, a paucity of immunological reagents has hampered the study of cellular immune responses. Here we describe the development and characterisation of a novel monoclonal antibody (mAb) against the ferret CD4 antigen and the characterisation of ferret CD4 T lymphocytes. | 3 |
| 2019 | Mice | Sarah R Leist | Increasing the translation of mouse models of MERS coronavirus pathogenesis through kinetic hematological analysis | We recently generated a mouse model for MERS-CoV pathogenesis through the humanization of the orthologous entry receptor dipeptidyl peptidase 4 (DPP4). To increase the translatability of this model, we validated and established the use of an automated veterinary hematology analyzer (VetScan HM5) at biosafety level 3 for analysis of peripheral blood. | 2 |
Capabilities to discover a therapeutic (not vaccine) for the disease, and clinical effectiveness studies to discover therapeutics, to include antiviral agents.¶
The majority of therapeutics under investigation consist of convalescent plasma and monoclonal/polyclonal antibodies that neutralize activity agains the virus spike protein.
An important finding suggests that there is one specific human monoclonal antibody (the epitope of CR3022) that does not overlap with the ACE2 binding site within 2019-nCoV RBD.
These results suggest that CR3022 may have the potential to be developed as candidate therapeutics, alone or in combination with other neutralizing antibodies, for the prevention and treatment of 2019-nCoV infections.
The same article suggests some differences between the RBD of SARS-CoV and 2019-nCoV.
Some of the most potent SARS-CoV-specific neutralizing antibodies (e.g. m396, CR3014) that target the ACE2 binding site of SARS-CoV failed to bind 2019-nCoV spike protein, implying that the difference in the RBD of SARS-CoV and 2019-nCoV has a critical impact for the cross-reactivity of neutralizing antibodies, and that it is still necessary to develop novel monoclonal antibodies that could bind specifically to 2019-nCoV RBD.
This relates to the findings of the 3D model solved by Jason S. McLellan that was mentioned earlier.

library(highcharter)
library(treemap)
library(viridis)
library(RColorBrewer)
therapeuticspapers <- read.csv("https://raw.githubusercontent.com/mdurazob/covidnotebook/master/therapeuticspapers.csv")
hchart(therapeuticspapers, "treemap", hcaes(x = Name, value = Citations, color = Year))%>%
hc_tooltip(pointFormat = "<b>Title: {point.Title}</b><br> <b>Keyword: {point.Name}</b><br> Fragment: {point.Abstract}") %>%
hc_colorAxis(minColor = brewer.pal(9, "Greens")[4],
maxColor = brewer.pal(9, "Blues")[9])
| Year | Name | Author | Title | Abstract | Citations | FIELD7 |
|---|---|---|---|---|---|---|
| 2004 | monoclonal antibody | Tong Zhou | An Exposed Domain in the Severe Acute Respiratory Syndrome Coronavirus Spike Protein Induces Neutralizing Antibodies | The properly refolded protein fragment specifically bound to the surface of Vero cells. Monoclonal antibodies raised against this fragment recognized the native spike protein of SARS CoV in both monomeric and trimeric forms. These monoclonal antibodies were capable of blocking S-II attachment to Vero cells and exhibited in vitro antiviral activity. | 73 | |
| 2007 | Polygonaceae herb | Ho TY | Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction | Emodin, an anthraquinone compound derived from genus Rheum and Polygonum, significantly blocked the S protein and ACE2 interaction in a dose-dependent manner. It also inhibited the infectivity of S protein-pseudotyped retrovirus to Vero E6 cells. These findings suggested that emodin may be considered as a potential lead therapeutic agent in the treatment of SARS. | 78 | |
| 2015 | 80R | J Virol | Evaluation of Human Monoclonal Antibody 80R for Immunoprophylaxis of Severe Acute Respiratory Syndrome by an Animal Study, Epitope Mapping, and Analysis of Spike Variants | In addition, the effects of various 80R-binding domain amino acid substitutions which occur in SARS-like-CoV from civet cats, and which evolved during the 2002/2003 outbreak and in a 2003/2004 Guangdong index patient, were analyzed. The results demonstrated that the vast majority of SARS-CoVs are sensitive to 80R. | 111 | |
| 2016 | griffithsin | Millet JK | Middle East respiratory syndrome coronavirus infection is inhibited by griffithsin | Coronaviruses are enveloped viruses, with the spike proteins present on their surface responsible for virus entry into the target cell. Lectins are attractive anti-coronavirus candidates because of the highly glycosylated nature of the spike protein. We tested the antiviral effect of griffithsin (GRFT), a lectin isolated from the red marine alga Griffithsia sp. against MERS-CoV infection. | 14 | |
| 2017 | MERS-five-helix bundle | Yaping Sun | Identification of a Novel Inhibitor against Middle East Respiratory Syndrome Coronavirus | The continual outbreaks of MERS-CoV highlight the importance of developing antiviral therapeutics. Here, we rationally designed a novel fusion inhibitor named MERS-five-helix bundle (MERS-5HB) derived from the six-helix bundle (MERS-6HB) which was formed by the process of membrane fusion. | 6 | |
| 2017 | monoclonal antibody | Du L | MERS-CoV spike protein: a key target for antivirals | This review illustrates MERS-CoV S proteins structure and function, particularly S1 receptor-binding domain (RBD) and S2 heptad repeat 1 (HR1) as therapeutic targets, and summarizes current advancement on developing anti-MERS-CoV therapeutics, focusing on neutralizing monoclonal antibodies (mAbs) and antiviral peptides. | 52 | |
| 2018 | peptide inhibitor | S Xia | Peptide-Based Membrane Fusion Inhibitors Targeting HCoV-229E Spike Protein HR1 and HR2 Domains | Here we reported the design, synthesis and characterization of two peptide-based membrane fusion inhibitors targeting HCoV-229E spike protein heptad repeat 1 (HR1) and heptad repeat 2 (HR2) domains, 229E-HR1P and 229E-HR2P, respectively. We found that 229E-HR1P and 229E-HR2P could interact to form a stable six-helix bundle and inhibit HCoV-229E spike protein-mediated cell-cell fusion with IC50 of 5.7 and 0.3 µM, respectively. | 17 | |
| 2018 | nanobody | Zhao G | A Novel Nanobody Targeting Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Receptor-Binding Domain Has Potent Cross-Neutralizing Activity and Protective Efficacy against MERS-CoV | Nanobodies (Nbs) are single-domain antibodies derived from camelids and sharks and are potentially cost-effective antivirals with small size and great expression yield. In this study, we developed a novel neutralizing Nb (NbMS10) and its human-Fc-fused version (NbMS10-Fc), both of which target the MERS-CoV spike protein receptor-binding domain (RBD). | 20 | |
| 2018 | monoclonal antibody | Hui-Ju Han | Neutralizing Monoclonal Antibodies as Promising Therapeutics against Middle East Respiratory Syndrome Coronavirus Infection | There are no approved vaccines or therapies for MERS until now. Passive immunotherapy with neutralizing monoclonal antibodies (mAbs) is an effective prophylactic and therapeutic reagent against emerging viruses. In this article, we review current advances in neutralizing mAbs against MERS-CoV. | 8 | |
| 2018 | IBV | Sisk JM | Coronavirus S protein-induced fusion is blocked prior to hemifusion by Abl kinase inhibitors | Here, we use IBV, a BSL-2 virus, as a model for studying the role of Abl kinase activity during coronavirus infection. We found that imatinib and two specific Abl kinase inhibitors, GNF2 and GNF5, reduce IBV titres by blocking the first round of virus infection. | 6 | |
| 2018 | TMPRSS2 | Hannah Kleine | Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein | These findings suggest that cleavage at S2? is carried out by proteases recognizing a single arginine, most likely TMPRSS2 and cathepsin L. | 9 | |
| 2018 | ?-Helical Peptides | Chao Wang | Discovery of Hydrocarbon-Stapled Short ?-Helical Peptides as Promising Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Fusion Inhibitors | The hexameric ?-helical coiled-coil formed between the C-terminal and N-terminal heptad repeat (CHR and NHR) regions of class I viral fusion proteins plays an important role in mediating the fusion of the viral and cellular membranes and provides a clear starting point for molecular mimicry that drives viral fusion inhibitor design. | 13 | |
| 2019 | m336 | Cong Wang | Combining a Fusion Inhibitory Peptide Targeting the MERS-CoV S2 Protein HR1 Domain and a Neutralizing Antibody Specific for the S1 Protein Receptor-Binding Domain (RBD) Showed Potent Synergism against Pseudotyped MERS-CoV with or without Mutations in RBD | However, we herein report that the combination of m336 and HR2P-M2 exhibited potent synergism in inhibiting MERS-CoV S protein-mediated cell–cell fusion and infection by MERS-CoV pseudoviruses with or without mutations in the RBD, resulting in the enhancement of antiviral activity in contrast to either one administered alone. | 8 | |
| 2019 | antibodies | Zhou Y | Advances in MERS-CoV Vaccines and Therapeutics Based on the Receptor-Binding Domain | The receptor-binding domain (RBD) of the S protein contains a critical neutralizing domain and is an important target for development of MERS vaccines and therapeutics. In this review, we describe the relevant features of the MERS-CoV S-protein RBD, summarize recent advances in the development of MERS-CoV RBD-based vaccines and therapeutic antibodies, and illustrate potential challenges and strategies to further improve their efficacy. | 9 | |
| 2019 | IgG antibody | Kim MH | Superior immune responses induced by intranasal immunization with recombinant adenovirus-based vaccine expressing full-length Spike protein of Middle East respiratory syndrome coronavirus | Here, we designed recombinant replication-deficient adenovirus-based vaccines expressing the N-terminal domain (rAd/NTD) and receptor-binding domain (rAd/RBD) of the MERS-CoV S1 subunit and full-length Spike protein (rAd/Spike). We found that immunization with candidate vaccines via intranasal route induced S1-specific IgG antibodies and neutralizing antibodies against MERS spike pseudotyped virus. | 3 | |
| 2019 | IBV | Jason Westerbeck | The Infectious Bronchitis Coronavirus Envelope Protein Alters Golgi pH To Protect the Spike Protein and Promote the Release of Infectious Virus | This is the first demonstration of a coronavirus-induced alteration in the microenvironment of the secretory pathway. | 9 | |
| 2020 | CR3022 | Tian X | Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody | Here, we report for the first time that a SARS-CoV-specific human monoclonal antibody, CR3022, could bind potently with 2019-nCoV RBD (KD of 6.3 nM). The epitope of CR3022 does not overlap with the ACE2 binding site within 2019-nCoV RBD. These results suggest that CR3022 may have the potential to be developed as candidate therapeutics, alone or in combination with other neutralizing antibodies, for the prevention and treatment of 2019-nCoV infections. | 44 |
Alternative models to aid decision makers in determining how to prioritize and distribute scarce, newly proven therapeutics as production ramps up. This could include identifying approaches for expanding production capacity to ensure equitable and timely distribution to populations in need.¶
Multiple research uncovers the budgetary restrictions and resource allocation problems that public policy can face during an epidemic.
A 2016 study identified multiple cost-effective policies to control epidemics, some of them relate to:
(i) when to employ or lift a transmission-reducing intervention (e.g. school closure) and (ii) how to prioritize population members for vaccination when a limited quantity of vaccines first become available
Another research paper used an stochastic epidemiological modelling approach to ellaborate cost-minimization strategies to contain the spread of epidemics in the population
library(highcharter)
economicspapers <- read.csv("https://raw.githubusercontent.com/mdurazob/covidnotebook/master/economicsPapers.csv")
economicschart <- economicspapers %>%
hchart('bubble', hcaes(x = 'Name', y = 'Year', size = 'Citations', color='Year')) %>%
hc_tooltip(pointFormat = "<b>Title: {point.Title}</b><br> <b>Keyword: {point.Name}</b><br> Fragment: {point.Abstract}")
economicschart
| Year | Name | Author | Title | Abstract | Citations |
|---|---|---|---|---|---|
| 2008 | critical care capacity | Rubinson L | Definitive Care for the Critically Ill During a Disaster: A Framework for Optimizing Critical Care Surge Capacity From a Task Force for Mass Critical Care Summit Meeting, January 26–27, 2007, Chicago, IL | Shifting critical care disaster preparedness efforts to augment limited, essential critical care (emergency mass critical care [EMCC]), rather than to marginally increase unrestricted, individual-focused critical care may provide many additional people with access to life-sustaining interventions. | 99 |
| 2010 | component-based | Jacco Wallinga | Optimizing infectious disease interventions during an emerging epidemic | Here we present first principles for allocating scarce resources with limited data. We show that under a broad class of assumptions, the simple rule of targeting intervention measures at the group with the highest risk of infection per individual will achieve the largest reduction in the transmission potential of a novel infection. | 123 |
| 2011 | Dynamic prediction | A Uribe | Predictive and Reactive Distribution of Vaccines and Antivirals during Cross-Regional Pandemic Outbreaks | We develop a large-scale simulation-driven optimization model for generating dynamic predictive distribution of vaccines and antivirals over a network of regional pandemic outbreaks. The model incorporates measures of morbidity, mortality, and social distancing, translated into the cost of lost productivity and medical expenses. | 0 |
| 2011 | surveillance economics | Barbara Hasler | Conceptualising the technical relationship of animal disease surveillance to intervention and mitigation as a basis for economic analysis | Surveillance and intervention are resource-using activities of strategies to mitigate the unwanted effects of disease. Resources are scarce, and allocating them to disease mitigation instead of other uses necessarily involves the loss of alternative sources of benefit to people. | 0 |
| 2012 | optimal vaccine coverage | Elodie Adida | Operational issues and network effects in vaccine markets | Although vaccines provide the most effective means for preventing infectious diseases, there are two main reasons why it is often difficult to reach a socially optimal level of vaccine coverage: (i) the emergence of operational issues (such as yield uncertainty) on the supply side, and (ii) the existence of negative network effects on the consumption side. | 33 |
| 2012 | stochastic modelling | Adam Kleczkowski | Searching for the most cost-effective strategy for controlling epidemics spreading on regular and small-world networks | The choice of the optimal strategy is governed mainly by a relative cost of palliative and preventive treatments. If the disease spreads within the well-defined neighbourhood, the local strategy is optimal unless the cost of a single vaccine is much higher than the cost associated with hospitalization. In the latter case, it is most cost-effective to refrain from prevention. | 35 |
| 2013 | public policy | JW Timbie | Systematic Review of Strategies to Manage and Allocate Scarce Resources during Mass Casualty Events | We analyzed published evidence on strategies to optimize the management and allocation of scarce resources across a wide range of MCE contexts and study designs. | 68 |
| 2016 | dynamic resource allocation | Reza Yaesoubi | Identifying Cost-Effective Dynamic Policies to Control Epidemics | We describe a mathematical decision model for identifying dynamic health policies for controlling epidemics. These dynamic policies aim to select the best current intervention based on accumulating epidemic data and the availability of resources at each decision point. We propose an algorithm to approximate dynamic policies that optimize the population’s net health benefit, a performance measure which accounts for both health and monetary outcomes. | 6 |
| 2017 | Pontryagin’s Minimum Principle | Luca Bolzoni | Time-optimal control strategies in SIR epidemic models | Applying the Pontryagin’s Minimum Principle (PMP) to the unconstrained control problems (i.e. without costs of control or resource limitations), we prove that, for all the policies investigated, only bang–bang controls with at most one switch are admitted. | 8 |
| 2018 | linear regression | Dimitrious Gouglas | Estimating the cost of vaccine development against epidemic infectious diseases: a cost minimisation study | We used a linear regression model to identify drivers of development costs from preclinical through to end of phase 2a. Drawing from published estimates of vaccine research and development probabilities of success, we simulated costs for advancing these 224 vaccine candidates through to the end of phase 2a. We combined these findings to determine minimum costs for progressing at least one vaccine through to the end of phase 2a per epidemic infectious disease by means of a stochastic optimisation model. | 20 |
| 2019 | Point-of-care testing | Kost GJ | Geospatial Science and Point-of-Care Testing: Creating Solutions for Population Access, Emergencies, Outbreaks, and Disasters | POCT-enabled spatial grids can map sentinel cases and establish geographic limits of epidemics for ring vaccination. | 0 |
| 2019 | Accumulated Error | KV Parag | Optimising Renewal Models for Real-Time Epidemic Prediction and Estimation | The effective reproduction number, Rt, is an important prognostic for infectious disease epidemics. Significant changes in Rt can forewarn about new transmissions or predict the efficacy of interventions. The renewal model infers Rt from incidence data and has been applied to Ebola virus disease and pandemic influenza outbreaks, among others. | 0 |
| 2020 | bayesian smoothing | Sarah McCough | Nowcasting by Bayesian Smoothing: A flexible, generalizable model for real-time epidemic tracking | Here, we present a Bayesian approach, NobBS (Nowcasting by Bayesian Smoothing) capable of producing smooth and accurate nowcasts in multiple disease settings. | 0 |
| 2020 | State-transition-matrix | Ke Wu Sr | Utilize State Transition Matrix Model to Predict the Novel Corona Virus Infection Peak and Patient Distribution | We used data resource according to cases reported by the National Health Commission of the Peoples Republic of China (Jan 25, 2019, to Feb 28, 2020) as the training set to deduce the arrival of the peak infection time and the number of severe and critical cases in Wuhan on subsequent days. | 0 |
| 2020 | exponential growth | Q Q Song | Study on assessing early epidemiological parameters of coronavirus disease epidemic in China | Considering the epidemic is ongoing, the exponential growth model was used to fit the incidence data of COVID-19 from 10 to 31 January, 2020, and exponential growth method, maximum likelihood method and SEIR model were used to estimate the basic reproduction number. Results: Early COVID-19 cases kept an increase in exponential growth manner before 26 January, 2020, then the increase trend became slower. | 0 |
Efforts targeted at a universal coronavirus vaccine.¶
Altough there is not much data about a universal coronavirus vaccine, there is one study that mentions the possibility of developing an epitope based vaccine against all strains of human coronaviruses. The main results were the following:
Multiple sequence alignment (MSA) approach was employed among spike (S), membrane (M), enveloped (E) and nucleocapsid (N) protein and replicase polyprotein 1ab to identify which one is highly conserve in all coronaviruses strains. Next, we use various in silico tools to predict consensus immunogenic and conserved peptide. We found that conserved region is present only in the RNA directed RNA polymerase protein. In this protein we identified one epitope WDYPKCDRA is highly immunogenic and 100% conserved among all available human coronavirus strains.
Efforts to develop animal models and standardize challenge studies¶
A very meaningful article published in the Center for Disease Control and Prevention highlighted the need to include standarization of animal models and virus stocks for studying disease pathogenesis and efficacy of medical countermeasures.
Use of standardized clinical data collection instruments and common biologic sampling protocols for serial prospective data collection will facilitate data pooling from MERS-CoV cases and comparisons across clinical sites and countries. Global collaborations among clinical networks are also needed to implement clinical trials, preferably randomized controlled clinical trials, of MERS-CoV investigational therapeutics,
There is an urgent need for an international agreement on the protocols and standarization of data collection methods.
The ISARIC-WHO Data tools for Severe Acute Respiratory is an effort to provide a from designed to collect data obtained through examination, interview, and review of hospital notes. An example of the form can be found here
library(highcharter)
library(treemap)
library(viridis)
library(RColorBrewer)
standarizationpapers <- read.csv("https://raw.githubusercontent.com/mdurazob/covidnotebook/master/AnimalStandardized.csv")
hchart(standarizationpapers, "treemap", hcaes(x = Name, value = Citations, color = Year))%>%
hc_tooltip(pointFormat = "<b>Title: {point.Title}</b><br> <b>Keyword: {point.Name}</b><br> Fragment: {point.Abstract}") %>%
hc_colorAxis(minColor = brewer.pal(9, "Greens")[4],
maxColor = brewer.pal(9, "Blues")[9])
| Year | Name | Author | Title | Abstract | Citations |
|---|---|---|---|---|---|
| 2006 | 180kDa viral spike protein | A Roberts | Animal models and antibody assays for evaluating candidate SARS vaccines: Summary of a technical meeting 25–26 August 2005, London, UK | In the absence of clinical cases of SARS, candidate vaccines should be evaluated for efficacy in animal models, and although it is uncertain whether the United States Food and Drug Administrations "animal rule" would apply to licensure of a SARS vaccine, it is important to develop standardized animal models and immunological assays in preparation for this eventuality. | 15 |
| 2008 | cotton rats | DM Watts | Evaluation of Cotton Rats as a Model for Severe Acute Respiratory Syndrome | Blood and turbinate wash samples, and lung tissue were collected from each animal at different time points after SARS coronavirus (CoV) infection for determining the growth curve of virus, if any, by the standard infectivity assay in Vero E6 cells. | 2 |
| 2016 | dromedaries | TM Uyeki | Development of Medical Countermeasures to Middle East Respiratory Syndrome Coronavirus | Research priorities include standardization of animal models and virus stocks for studying disease pathogenesis and efficacy of medical countermeasures; development of MERS-CoV diagnostics; improved access to nonhuman primates to support preclinical research; studies to better understand and control MERS-CoV disease, including vaccination studies in camels; and development of a standardized clinical trial protocol. | 12 |
| 2019 | rabbit | AS Macedo | Animal modeling in bone research—Should we follow the White Rabbit? | In that sense, standardizing novel models remains necessary and rabbits are gaining in popularity as potential bone models. | 0 |
Efforts to develop prophylaxis clinical studies and prioritize in healthcare workers¶
There are some studies that assess the efficacy of certain drugs as Post Exposure Prophylaxis (PEP) strategies for healthcare workers.
Two studies (one made in 2018 and another in 2019) assessed the efficacy of ribavirin and lopinavir/ritonavir as PEP therapy.
A more recent study made in 2020 during the Wuhan breakout established a treatment strategy centered on "Four Anti and Two-Balance" which include anti-virus, anti-shock, anti-hyoxemia,anti-secondary infection, and maintaining of water, electrolyte and acid base balance and microecological balance. It is said that the "Four-Anti and Two-Balance" strategy effectively increased cure rate and reduced mortality.
library(highcharter)
prophylaxispapers <- read.csv("https://raw.githubusercontent.com/mdurazob/covidnotebook/master/prophylaxispapers.csv")
prophylaxischart <- prophylaxispapers %>%
hchart('bubble', hcaes(x = 'Name', y = 'Year', size = 'Citations', color='Year')) %>%
hc_tooltip(pointFormat = "<b>Title: {point.Title}</b><br> <b>Keyword: {point.Name}</b><br> Fragment: {point.Abstract}")
prophylaxischart
| Year | Name | Author | Title | Abstract | Citations |
|---|---|---|---|---|---|
| 2002 | Chemoprophylaxis | Semin Perinatol | Hospital-acquired viral pathogens in the neonatal intensive care unit | The intensive care environment allows for efficient spread of viral pathogens, and secondary cases among both patients and healthcare workers are frequently observed | 42 |
| 2006 | Blood/body fluids | Tarantola A | Infection risks following accidental exposure to blood or body fluids in health care workers: A review of pathogens transmitted in published cases | Although their principal task remains the evaluation of the risks of HIV, HBV, or HCV transmission and the possible usefulness of postexposure prophylaxis, these experts are also responsible for evaluating risks of occupational infection with other emergent or more rare pathogens and their possible timely prevention | 338 |
| 2007 | targeted antiviral | MG Roberts | A model for the spread and control of pandemic influenza in an isolated geographical region | The model is then modified to reflect a variety of control strategies based on social distancing measures, targeted antiviral treatment and antiviral prophylaxis and home quarantine, and the effectiveness of the strategies is compared. | 58 |
| 2016 | teicoplanin | J Biol | Glycopeptide Antibiotics Potently Inhibit Cathepsin L in the Late Endosome/Lysosome and Block the Entry of Ebola Virus, Middle East Respiratory Syndrome Coronavirus (MERS-CoV), and Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) | Furthermore, teicoplanin also has an inhibitory effect on transcription- and replication-competent virus-like particles, with an IC50 as low as 330 nm Comparative analysis further demonstrated that teicoplanin is able to block the entry of Middle East respiratory syndrome (MERS) and severe acute respiratory syndrome (SARS) envelope pseudotyped viruses as well | 42 |
| 2018 | ribavirin/lopinavir | So Yeon Park | 2491. Post-Exposure Prophylaxis With Ribavirin Plus Lopinavir/Ritonavir for Middle East Respiratory Syndrome in Healthcare Workers | In this study, we assessed (1) the efficacy of oral ribavirin and lopinavir/ritonavir as PEP for HCWs exposed to patients with severe MERS-CoV pre-isolation pneumonia, and (2) safety of the PEP regimen. | 0 |
| 2019 | ribavirin/lopinavir | SY Park | Post-exposure prophylaxis for Middle East respiratory syndrome in healthcare workers | This study assessed the efficacy of ribavirin and lopinavir/ritonavir as PEP for healthcare workers (HCWs) exposed to patients with severe MERS-CoV pre-isolation pneumonia. The safety of the PEP regimen was assessed. HCWs with high-risk exposure to MERS-CoV pre-isolation pneumonia were retrospectively enrolled. HCWs who received PEP therapy were classified into the PEP group. PEP therapy was associated with a 40% decrease in the risk of infection. There were no severe adverse events during PEP therapy. | 10 |
| 2020 | Four-Anti and Two-Balance | Kaijin Xu | Management of corona virus disease-19 (COVID-19): the Zhejiang experience | The "Four-Anti and Two-Balance"strategy included antivirus, anti-shock, anti-hyoxemia, anti-secondary infection, and maintaining of water, electrolyte and acid base balance and microecological balance. Meanwhile, integrated multidisciplinarypersonalized treatment was recommended to improve therapeutic effect | 15 |
Approaches to evaluate risk for enhanced disease after vaccination¶
A recurrent theme is the need to evaluate immune responses after vaccination.
A 2019 research dealing with the immunogenicity of different forms of MERS S Glycoprotein found significant immune responses after vaccination with the full length S (rAd5-S).
A 2018 article found T Cells responding to the MERS-CoV nucleocapsid protein delivered by Vaccinia Virus MVA in Mice.
An important part of the studies found different responses in the T cell after vaccination. More specifically one research article published in 2020 found that a soluble version of Nipah Virus Glycoprotein G delivered by vaccinia virus MVA activates Specific CD8 and CD4 T Cells in mice. Altough this study does not deal with MERS-CoV, valuable knowledge can be extracted from this findings.
library(highcharter)
library(treemap)
library(viridis)
library(RColorBrewer)
aftervaccinepapers <- read.csv("https://raw.githubusercontent.com/mdurazob/covidnotebook/master/aftervaccinepapers.csv")
hchart(aftervaccinepapers, "treemap", hcaes(x = Name, value = Citations, color = Year))%>%
hc_tooltip(pointFormat = "<b>Title: {point.Title}</b><br> <b>Keyword: {point.Name}</b><br> Fragment: {point.Abstract}") %>%
hc_colorAxis(minColor = brewer.pal(9, "Greens")[4],
maxColor = brewer.pal(9, "Blues")[9])
| Year | Name | Author | Title | Abstract | Citations |
|---|---|---|---|---|---|
| 2019 | V920 | Monath TP | rVSV?G-ZEBOV-GP (also designated V920) recombinant vesicular stomatitis virus pseudotyped with Ebola Zaire Glycoprotein: Standardized template with key considerations for a risk/benefit assessment | During the first few days after vaccination, many vaccinees experience a mild acute-phase reaction with fever, headache, myalgia, and arthralgia of short duration; this period is associated with a low-level viremia, activation of anti-viral genes, and increased levels of chemokines and cytokines. Oligoarthritis and rash appearing in the second week occur at a low incidence, and are typically mild-moderate in severity and self-limited. | 9 |
| 2018 | MVvac2-MERS-S(H) | BS Bodmer | Live-attenuated bivalent measles virus-derived vaccines targeting Middle East respiratory syndrome coronavirus induce robust and multifunctional T cell responses against both viruses in an appropriate mouse model | MVvac2-MERS-S(H) induces robust humoral and cellular immunity against MERS-S mediating protection. Here, the induction and nature of immunity after vaccination with MVvac2-MERS-S(H) or novel MVvac2-MERS-N were further characterized. We focused on the necessity for vector replication and the nature of induced T cells, since functional CD8+ T cells contribute importantly to clearance of MERS-CoV. | 8 |
| 2018 | alum+CpG | Y Deng | Enhanced protection in mice induced by immunization with inactivated whole viruses compare to spike protein of middle east respiratory syndrome coronavirus | No antigen-specific T-cell immunity was detected after vaccination based on the interferon-? ELISpot assay. Mice were transduced with Ad5-hDPP4 after the final immunization and were then challenged with MERS-CoV (1?×?105 plaque-forming units). Compared with the control group (adjuvant alone), mice immunized with the S protein or IV showed slightly lower pathological damage in the lung, as well as reduced antigen expression and lung virus titers. | 6 |
| 2020 | MVA–NiV-G | Georgia Kalodimou | A Soluble Version of Nipah Virus Glycoprotein G Delivered by Vaccinia Virus MVA Activates Specific CD8 and CD4 T Cells in Mice | Overlapping peptides covering the entire NiV-G protein were used to identify major histocompatibility complex class I/II-restricted T cell responses in type I interferon receptor-deficient (IFNAR?/?) mice after vaccination with the MVA–NiV-G candidate vaccines | 0 |
| 2018 | T cell response | Roberts LM | Temporal requirement for pulmonary resident and circulating T cells during virulent Francisella tularensis infection | Despite the dominant in vivo response by parenchymal CD4+ T cells after vaccination and challenge, circulating CD4+ T cells were superior at controlling intracellular Ftt replication in vitro. Further examination in vivo revealed temporal requirements for resident and circulating T cells during Ftt infection. | 1 |
| 2017 | T cell response | Allison Chirstiansen | CD11a and CD49d enhance the detection of antigen-specific T cells following human vaccination | A phase I vaccine trial enabled us to evaluate a novel gating strategy based on surface expression of CD11a and CD49d as a means of detecting antigen-specific, cytokine producing CD4 and CD8 T cells induced after vaccination of naïve individuals against leishmaniasis. | 5 |
| 2012 | T cell response | Shih-Min Wang | The regulatory T cells in anti-influenza antibody response post influenza vaccination | The plasma level of the transforming growth factor (TGF-?), but not interleukin (IL)-2, IL-4, IL-5, IL-10, IFN-?, TNF-?, was also found to increase significantly after vaccination. | 25 |
| 2019 | rAd5-S | Ozharovskaia TA | Immunogenicity of Different Forms of Middle East Respiratory Syndrome S Glycoprotein | The most significant cellular immune response was induced after vaccination of the animals with the full-length S (rAd5-S). These investigations suggest that the full-length S and the membrane form of the RBD (RBD-G) are the most promising vaccine candidates among all the studied forms of S glycoprotein. | 1 |
| 2018 | Ad5-hDPP4 | Deng Y | Enhanced protection in mice induced by immunization with inactivated whole viruses compare to spike protein of middle east respiratory syndrome coronavirus | No antigen-specific T-cell immunity was detected after vaccination based on the interferon-? ELISpot assay. Mice were transduced with Ad5-hDPP4 after the final immunization and were then challenged with MERS-CoV (1?×?105 plaque-forming units). | 6 |
| 2018 | T cell response | Veit S | CD8+ T Cells Responding to the Middle East Respiratory Syndrome Coronavirus Nucleocapsid Protein Delivered by Vaccinia Virus MVA in Mice. | Antibodies directed against the MERS-CoV spike (S) protein are thought to play a major role in controlling MERS-CoV infection and in mediating vaccine-induced protective immunity. In contrast, relatively little is known about the role of T cell responses and the antigenic targets of MERS-CoV that are recognized by CD8+ T cells. | 7 |
Assays to evaluate vaccine immune response and process development for vaccines, alongside suitable animal models [in conjunction with therapeutics]¶
The summary of a 2005 technical meeting in London mentioned earlier summarizes the animal models and antibody assays for evaluating candidate SARS vaccines, the study highlights that:
In selecting animal models for vaccine evaluation, it is important to remember the principle underlying the so called “animal rule”, where data from more than one animal species is often required: each animal species should contribute something different to our understanding of disease and protection.
Upon this guideline the recommended animal models include Inbred mouse strains (BALB/c, C57BL/6, 129SvEv, STAT1−/−) whom support SARS-CoV replication and can develop pneumonitis (129S). Golden Syrian hamsters are also an excellent animal model because they demonstrate high levels of SARS-CoV replication and develop pneumonitis. Hamsters are suitable for vaccine efficacy, immunoprophylaxis and treatment studies.
Among non-human primates (NHP) are the common marmoset and African green monkeys (AGMs, Chlorocebus aethiops sabeus).
The SARSCoV strains tested in NHP models are HKU-39849 (cynos), PUMC (rhesus) and Urbani (marmosets and AGMs). These were inoculated by the respiratory route (IN, IT) at a dose of 106pfu or more/NHP.
Other studies provided the following findings:
In a 2019 study about Receptor-Binding domains, vaccine candidates RBD-TcdA, RBD-TcdB, and RBD-TcdA/B were evaluated by ELISA and survival assays performed on mice with RBD-TcdA and/or RBD-TcdB
In another study ELISPOT assay is used to determine T cell responses from 128 SARS convalescent samples.
A more recent study used the ferret model with an emphasis in human viral infectious diseases.
Another one highlights the importance of carefully assessing cytotoxicity in drug screen assays in primary human monocyte-derived macrophages (MDMs).
library(highcharter)
assaypapers <- read.csv("https://raw.githubusercontent.com/mdurazob/covidnotebook/master/assaypapers.csv")
assaychart <- assaypapers %>%
hchart('bubble', hcaes(x = 'Name', y = 'Year', size = 'Citations', color='Year')) %>%
hc_tooltip(pointFormat = "<b>Title: {point.Title}</b><br> <b>Keyword: {point.Name}</b><br> Fragment: {point.Abstract}")
assaychart
| Year | Name | Author | Title | Abstract | Citations |
|---|---|---|---|---|---|
| 1998 | Cytotoxicity | L Pewe | Infection with Cytotoxic T-Lymphocyte Escape Mutants Results in Increased Mortality and Growth Retardation in Mice Infected with a Neurotropic Coronavirus | These mutations abrogated recognition by T cells harvested from the central nervous systems of infected mice in direct ex vivo cytotoxicity assays. | 35 |
| 2001 | Immunospot | Yuan L | Protective Immunity and Antibody-Secreting Cell Responses Elicited by Combined Oral Attenuated Wa Human Rotavirus and Intranasal Wa 2/6-VLPs with Mutant Escherichia coli Heat-Labile Toxin in Gnotobiotic Pigs | The isotype, magnitude, and tissue distribution of antibody-secreting cells (ASCs) in the intestinal and systemic lymphoid tissues were evaluated using an enzyme-linked immunospot assay. | 65 |
| 2002 | Lymphoproliferative | Arriba ML | Lymphoproliferative responses and protection in conventional piglets inoculated orally with virulent or attenuated porcine epidemic diarrhoea virus | A lymphoproliferative assay was developed in which mononuclear cells isolated from lymphoid tissues at different postinoculation and postchallenge days underwent a secondary in vitro stimulation with semipurified antigen obtained from PEDV-infected cell cultures. | 22 |
| 2005 | Western blot | Zhang CH | Immune responses in Balb/c mice induced by a candidate SARS-CoV inactivated vaccine prepared from F69 strain | Western blot assay further demonstrated the specificity of the induced serum antibodies. | 20 |
| 2005 | Immunological | Roberts A | Animal models and antibody assays for evaluating candidate SARS vaccines: Summary of a technical meeting August 2005, London, UK | Administrations "animal rule" would apply to licensure of a SARS vaccine, it is important to develop standardized animal models and immunological assays in preparation for this eventuality.� | 15 |
| 2006 | Cytotoxicity | Tayade C | l-Arginine stimulates intestinal intraepithelial lymphocyte functions and immune response in chickens orally immunized with live intermediate plus strain of infectious bursal disease vaccine | Functional activities of iIELs evaluated by cytotoxicity assay demonstrated significantly high percentage cytotoxicity in IP+l-arginine groups compared with IP group (P<0.05). | 29 |
| 2008 | ICS/ELISpot/ELISA | Martin JE | A SARS DNA vaccine induces neutralizing antibody and cellular immune responses in healthy adults in a Phase I clinical trial | A single-plasmid DNA vaccine encoding the Spike (S) glycoprotein was evaluated in 10 healthy adults. Nine subjects completed the 3 dose vaccination schedule and were evaluated for vaccine safety and immune responses. Immune response was assessed by intracellular cytokine staining (ICS), ELISpot, ELISA, and neutralization assays. | 87 |
| 2008 | ELISPOT | Chris Ka-fai | T Cell Responses to Whole SARS Coronavirus in Humans | We used a matrix of overlapping peptides spanning whole SARS-CoV proteome to determine T cell responses from 128 SARS convalescent samples by ex vivo IFN-? ELISPOT assays. | 54 |
| 2012 | Immunosorbent | Cong H | Toxoplasma gondii HLA-B0702 restricted GRA7 peptide with adjuvants and an universal helper T cell epitope elicits CD8(+) T cells producing IFN-? and reduces parasite burden in HLA-B0702 mice | Herein, these peptides were evaluated to determine whether they could elicit IFN-? in splenocytes of HLA-B*0702 transgenic mice when administered with adjuvants and protect against subsequent challenge. Peptide-specific IFN-?-producing T cells were identified by enzyme-linked immunosorbent spot and proliferation assays | 37 |
| 2012 | ELISPOT | Papaneri | Further characterization of the immune response in mice to inactivated and live rabies vaccines expressing Ebola virus glycoprotein | Here, we expand our investigation of the immunogenic properties of these bivalent vaccines in mice. Both live and killed vaccines induced primary EBOV GP-specific T-cells and a robust recall response as measured by interferon-? ELISPOT assay. | 28 |
| 2015 | MxA /CRP point-of-care | Robert Sambursky and Nathan Shapiro | Evaluation of a combined MxA and CRP point-of-care immunoassay to identify viral and/or bacterial immune response in patients with acute febrile respiratory infection | A prospective, single center, blinded, observational clinical trial was conducted at Beth Israel Deaconess Medical Center from December 2012 to August 2013 to determine the accuracy of a POC immunoassay to identify a clinically significant immune response to viral and/or bacterial infection. | 21 |
| 2015 | In vitro toll | Vianka KF | Setting the proportion of CD4+ and CD8+ T-cells co-cultured with canine macrophages infected with Leishmania chagasi | The present study intended to establish an in vitro toll to assess the parasitological/immunological status of dogs, applicable in pre-clinical trials of vaccinology, prognosis follow-up and therapeutics analysis of canine visceral leishmaniasis.� | 6 |
| 2018 | Drug screen | Y Cong | MERS-CoV pathogenesis and antiviral efficacy of licensed drugs in human monocyte-derived antigen-presenting cells | These results demonstrate the value of testing drugs in primary cells, in addition to established cell lines, before initiating preclinical or clinical studies for MERS treatment and the importance of carefully assessing cytotoxicity in drug screen assays. | 10 |
| 2019 | ELISA/Survival | Luo D et al | Immunogenicity and Protection from Receptor-Binding Domains of Toxins as Potential Vaccine Candidates for Clostridium difficile | Vaccine candidates RBD-TcdA, RBD-TcdB, and RBD-TcdA/B was evaluated by ELISA and survival assays. The data indicated that mice immunized with all vaccine candidates displayed potent levels of RBD-specific serum IgG. | 0 |
| 2019 | ELISA | Xunliang Li | Porcine IL-12 plasmid as an adjuvant improves the cellular and humoral immune responses of DNA vaccine targeting transmissible gastroenteritis virus spike gene in a mouse model | The titers of TGEV S1 in an enzyme-linked immunosorbent assay (ELISA) and TGEV neutralizing antibodies and the concentrations of interferon (IFN)-? and IL-4 were also analyzed in the serum. | 1 |
| 2019 | Ferret | J Wong | Improving immunological insights into the ferret model of human viral infectious disease | Nevertheless, progress has been made to understand immune responses in the ferret model with a limited set of ferret?specific reagents and assays. | 0 |
Keyword Explorer and final words¶
By using the gexf file it was possible to build a keyword explorer using Gephi Gexf Viewer. This explorer lets the user search a terms and look at their connections. The layout was made using ForceAtlas 2 algorithm. This was the result:
Taking a network approach may be a simple matter compared to more complex machine learning methods. However, by using basic centrality and weight indicators the user can gain valuable knowledge about a particular field when coupled with the appropiate data visualizations. This approach can be used to examine the scientific literature on multiple topics. The insights provided by a network approach coupled with bibliometric management can throw light on emerging fields of research. Any comments on how to enhance this approach will be well received.